Nonstructural protein gene based molecular characterization of an attenuated goose parvovirus D strain.
Shao Wang1,2 , Xiaoli Zhu1,2 , Xiaoxia Cheng1,2 , Shifeng Xiao1,2 , Shaoying Chen1,2* , Fengqiang Lin1,2 , Shilong Chen1,2
1. Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agriculture Sciences, Fuzhou 350003, China
2. Fujian Animal Diseases Control Technology Development Center, Fuzhou 350013, China
Corresponding author:
Shaoying Chen, Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agriculture Sciences, No. 247 Wusi Road, Gulou District, Fuzhou 350003, China
Received: September 15, 2021
Accepted: September 22, 2021
Published: September 25, 2021
1. Abstract
1.1. To characterize the non-structural (NS) protein gene of the vaccine of muscovy duck (Cairina moschata) origin Goose parvovirus (GPV) D strain, a pair of primers were designed by DNAStar software based on the published sequences of the MDGPV-PT strain published in GenBank. The homology, evolution of heredity, glycosylation sites, phosphorylation sites, B-cell epitopes, T-cell epitopes and secondary structure of the NS protein were analyzed analyzedusing using bioinformatics software. The full-length NS gene was amplified using PCR and cloned into a pMD18-T vector. Sequencing analysis demonstrated that the NS gene of the MDGPV-D strain comprised 1884 bp encoding 627 amino acids. The MDGPV-Dstrain NS gene had considerable similarity to that of MDPV (nt: 97.9-98.6%, aa: 97.6-98.2%). Three potential N-glycosylation sites and 27 phosphorylation sites were found to possibly exist in the MDGPV-D strain NS protein, possibly containing 11 B-cell epitopes, 13 CD8+ CTL epitopes and 10 CD4+ Th epitopes. Prediction of the secondary structure of the NS protein revealed that the alpha helix, random coil and beta angle accounted for 40.67%, 41.15% and 4.63%, of the protein, respectively. Compared with the parental virulent PT strain, the attenuated vaccine strain D revealed that during the attenuation processes one common nucleotide change arose representing an amino acid substitution. The NS protein contained two amino acid substitutions, one being in the nucleoside triphosphate (NTP) binding motif at amino acid 338. The results of the present study provide a molecular basis for investigating attenuation mechanisms of MDGPV, and a useful reference for further studies of genetically engineered MDGPV vaccines.
1.2. Keywords: MDGPV; NS protein; Epitope; Molecular characteristics.
Introduction
Gosling plague is caused by the goose parvovirus (GPV) in goslings and young Muscovy ducks. The disease first occurred in 1956 in Jiangsu province, and soon spread to other habitats of the goose in China but was rarely observed in Muscovy ducks. Fang et al. established that the disease was caused by the goose parvovirus [1]. An outbreak of Muscovy duck gosling plague occurred in the Muscovyduck breeding regions of Putian and Fuqing in Fujian Province in 1997. It has now become endemic to all duck breeding regions, nationwide. Clinically, the disease is characterized by diarrhea and the formation of intestinal mucosal emboli in young Muscovy ducks. The incidence of the disease ranges from 50 to 70% and the fatality rate ranges from 40 to 65%, causing great economic losses to the Muscovy duck breeding industry.The pathogen was isolated and identified as a gosling plague virus. A weak strain with good safety and strong immunogenicity (MDGPV-D) was bred and used to develop a live vaccine to prevent the disease [2,3].
GPV is a member of the parvovirus family of viruses, the parvoviridae. The genome of the virus is a single-stranded linear DNA, with physicochemical characteristics consistent with the Muscovy duck parvovirus (MDPV). The genome is approximately 5000 bp in size and composed mainly of a 5’ non-structural (NS) protein gene and 3’ viral capsid protein VP gene [4,5]. NS proteins inhibit DNA replication in host cells and are associated with virulence, playing a key role in viral replication [6-8]. In the present study, the nucleotide sequence of the NS gene of the MDGPV-D strain was determined and the amino acid sequence of its NS protein derived. Bioinformatics analysis was performed using bioinformatics software, laying a foundation for further study of the correlation between genetic variation and change in virulence of the MDGPV-D strain.
Materials and methods
Virus strains
The weak MDGPV strain (MDGPV-D) was attenuated by the passage of the strong parent strain (MDGPV-PT), which was identified and preserved in the animal virus laboratory.
Primer design and PCR amplification of the NS gene
Total The NS gene sequence of the MDGPV-PT strain was extracted from GenBank, and a pair of primers designed with Oligo 6 software. The specific sense and antisense primers of the amplified complete ORF (1844bp) fragment of NS gene were synthesized by Bao-bioengineering Company (Dalian), as follows:
NSF: 5’-GGTCGGAGAGATGGCACTTTCT-3’,
NSR: 5’-ATTTTGAATCATCTTTATTGTTCAT-3’.
The viral DNA was extracted in accordance with the Qiagen instructions using a blood, tissue and cell DNA extraction kit from 200 μL of solution. In the present study, a 50 μL volume was analyzed, comprising prepared viral DNA 5 μL as template, upstream and downstream primers (20 pmol/L) 1 μL each, 2.5 mmol/L dNTPs 1 μL, 10×PCR Buffer 5 μL, 20 mmol/L MgCl2 0.8 μL, Easy-A high-fidelity DNA polymerase 0.2 μL, and sterilized water 36 μL. The amplification procedure was:pre- denaturation at 95 oC for 5 min, denaturation at 94 oC for 1 min, annealing at 52 oC for 1 min, extension at 72 oC for 2 min, for a total of 35 cycles, and a final extension at 72 oC for 10 min. PCR products were analyzed using 1% agarose gel electrophoresis which was used to observe the PCR amplification results.
Nucleotide analysis of the NS gene and its derived amino acid sequence
The Clustal W method was used to compare the nucleotide and derived amino acid sequences of the NS gene in the MDGPV-D strain with the corresponding sequences of 11 MDGPV, GPV and MDPV reference strains published in GenBank using the Megalign application in DNAStar Lasergene 7.1 software. Furthermore, conserved region analysis was conducted on the amino acid sequences derived from the Adeno-associated virus (AAV-2) strain of the NS gene. The 11 GPV and MDPV reference strains used in the present study were the GPV DY strain (Accession No.EF515837), GPV 82-0321v strain (Accession No. EU583389), GPV 82-0321 strain (Accession No.EU583390), GPV 06-0329 strain (Accession No.EU583391), GPV VG32/1 strain (Accession No.EU583392), GPV YG strain (Accession No.AF416726), GPV B strain (Accession No.U25749), MDGPV PT strain (Accession No.JF926695), MDPV FM strain (Accession No.NC_006147), MDPV P strain (Accession No.JF926697), MDPV P1vstrain(Accession No.JF926698) and Adenovirus AAV-2 strain (Accession No. NC_001401).
PhylogeneticTree analysis of the NS gene in MDGPV-D strain
The phylogenetic tree algorithm in the Megalign application was used to compare the NS gene sequence of MDGPV-D strain with the corresponding NS gene sequences of the MDGPV, GPV and MDPV reference strains listed above to construct phylogenetic trees.
Prediction of glycosylation and phosphorylation sites
of the NS protein in MDGPV-D strain
Glycosylation site prediction was conducted using NetNGlyc software (http://www.cbs.dtu.dk/services/NetNGlyc). Phosphorylation site prediction was conducted using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/).
Analysis of hydrophilicity, region flexibility, possibile surfaces and antigen index of NS protein in MDGPV-D strain, secondary structure prediction and predicted B-cell antigen epitopes
Analysis of the NS protein in the MDGPV-D strain was performed using the Protean application in DNAStar Lasergene 7.1 software to analyze hydrophilicity, regions of flexibility, macromolecular structure, motion and function. The SOPMA (http://www.expasy.ch/tools) protein structure prediction server was used to predict the secondary structure of the NS protein from the MDGPV D strain sequence. The results were analyzed to eliminate sequences in the secondary structures unable to form α-helical and β-folded epitopes. In addition, B-cell epitopes were predicted using the Bcepred prediction server (http://www.imtech.res.in/raghava/bcepred). Combined with the predicted results of the secondary structures, fragments with good plasticity and strong antigenicity were selected as candidate epitopes for further analysis.
Prediction of T-cell antigen epitopes of NS proteins in the MDGPV-D strain
MHC class-I and II restrictive T-cell binding regions in antigen epitopes were predicted using nHLAPredProPred software (http://www.imtech.res.in/) and mutationsinMCH I CD8+ cytotoxic T-cell epitopes and MCH II CD4+ T-cell epitopes compared between MDGPV-PT and D strains.
Results
Clone identification and sequencing of NS gene in MDGPV-D strain
The amplified sequence of the NS gene in the MDGPV-D strain and from recombinant plasmids demonstrated the presence of a1.8kb DNA fragment in both. After recovery and purification, the bands were ligated to a pMD18-Tvector, transformed into Escherichia coli. DH5α, and screened for positive clones. Bidirectional sequencing was performed on positive recombinant plasmids byBao-bioengineering Company (Dalian). Sequencing results indicated that the NS gene of the MDGPV-D strain was 1884 bp in length, including a complete open reading frame (ORF).
Prediction of glycosylation and phosphorylation sites of NS protein in MDGPV-D strain
Using the online software NetNGlyc 1.0, the NS protein in the MDGPV-D strain was predicted to contain three N-glycosylation sites: 150-NKTV, 225-NYSN and 360- NWTN. Online software NetPhos 2.0 predicted the following phosphorylation sites for the NS protein: 17 serine sites (11, 23, 117, 122, 200, 240, 251, 254, 391, 422, 434, 453, 464, 529, 542,
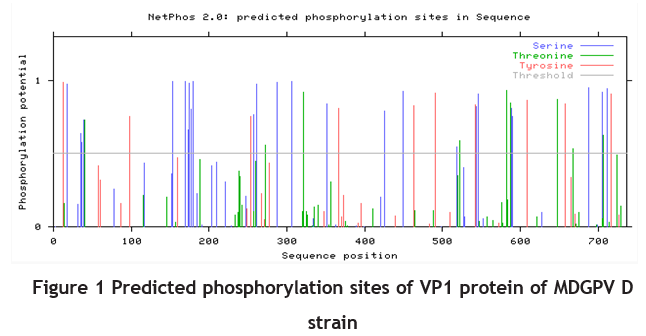
546, 587), 6 threoninesites (144, 339, 340, 385, 518, 552), and 4 tyrosinesites (95, 252, 285, 313) (Figure 1).
Predicted B-cell antigen epitopes in NS protein
The SOPMA server revealed that the secondary structure of NS protein in the MDGPV-D strain consisted of the following characteristics: alpha helix with 133 amino acids (aa), extended strand with 138 aa, beta turns with 33 aa and random coils with 428 aa, accounting for 18.17%, 18.85%, 4.51% and 58.47% of the protein, respectively. Hydrophilicity, flexible regions, probability of surfaces and antigen index were analyzed. The Bcepred B-cell epitope prediction server was used for analysis. The results indicate that the segments (aa 101-107, aa 139-105, aa 161-167, aa 336-342, aa 379-383, aa 401-408, aa 428-434, aa 458-464, aa 487-493, aa 502-510, aa 537-548) may be the dominant B-cell epitopes of the NS protein in the MDGPV-D strain
Predicted T-cell antigen epitopes in NS protein
T-cell epitopes can be divided into MCH I CD8+ CLT epitopes and MCH II CD4+T Th epitopes. nHLAPred and ProPred software predicted CTL epitopes based on the characteristics of the anchor residues of different HLA molecules. Thirteen peptide regions were predicted to be HLA-A2 specific antigen epitopes, namely: aa39-47, aa55-63, aa151-170, aa174-182, aa209-217, aa225-233, aa278-292, aa331-341, aa348-356, aa388-403, aa474-482, aa512-522, aa524-539. Th epitope peptides were predicted to be located in the sequence as follows:aa448-456, aa252-260, aa95-103, aa232-240, aa301- 309, aa569-577, aa179-187, aa395-403, aa572-580, and aa87- 95.
Homology analysis of NS gene in MDGPV-D strain
SeqMan analysis software was used to analyze the sequencing results of the NS gene (GenBank Accession No.JF926696) in the MDGPV-D strain. The Megalign application in DNAStar Lasergene 7.1 software was used to compare the 12 NS gene sequences from MDGPV, GPV and MDPV strains. There was no variation in terms of base deletion or insertion. Homologies of nucleotides and amino acid sequences in the D and PT strains were 99.6% and 98.9%, respectively. The corresponding homologies between the D strain and GPV reference strain were 79.7-81.8% and 90.0-90.8%, respectively, and 98.4-99.0% and 97.9-98.4% for homologies between the D strain and MDPV reference strain, respectively (Table 1).
Phylogenetic analysis of NS gene in MDGPV D strain
A phylogenetic tree of the NS gene in the MDGPV strain D and 11 other MDGPV, GPV and MDPV reference strains was plotted using MegAlign bioanalysis software. Based on gene sequence comparisons and phylogenetic tree analysis, although the complete nucleotide sequence and derived amino acid sequence of the MDGPV NS gene was close to the evolutionary relationship of MDPV, the first 36 nucleotides (nt1-nt36) and the corresponding 12 amino acids (aa1-aa12) in the open reading frame had clear GPV gene characteristics. Even if the nt1-nt195 segment of the MDGPV NS gene possessed MDPV gene sequence characteristics, its derived amino acid sequence (aa1-aa65) still genetically derived from the GPV NS protein (Figure 2A, 2B and 2C). One site where the aa sequence differed was found in the



MDGPV PT strain, aa338(A→V), compared to the D strain in the NTP region (aa336-aa343) (Figure 3).
Discussion
The MDGPV-D strain is the first successful cell-passage attenuated vaccine in China. The NS protein is the principal non-structural protein in GPV and plays a positive regulatory role in the activation of the capsid protein gene. Amplification sequencing and bioinformatics analysis of the NS protein gene in the MDGPV strain D has been used to study the molecular mechanisms of MDGPV attenuation and enrichment of \ molecular biological data.
In the present study, a Qiagen blood and tissue DNA extraction kit was used to isolate viral DNA, ensuring the quality of the single-stranded DNA template in samples. The target gene fragment was amplified using a high-fidelity PCR cloning enzymefrom Stratagene, which prevented degradation of primers and templates during the construction of the PCR reaction system, providing high-sensitivity and specific PCR reaction conditions for NS gene amplification of the MDGPV. Nucleotide sequence homology between the MDGPV-D strain NS gene and 11 MDGPV, GPV and MDPV NS genes from different hosts and different degrees of virulence recorded on GenBank in two regions of Europe and Asia, was 79.7%- 99.2%, while homology of the derived amino acid sequence was 90.0%-98.7%. According to the phylogenetic tree, the parental PT virulent strain of the NS gene in the MDGPV-D strain had considerable similarity to that of MDPV. However, its nucleotide and derived amino acid N-terminal sequences still retained GPV gene characteristics, suggesting that the genomes of members of the genus parvovirus are prone to recombination, consistent with the study of Shackelton et al [9-14]. MDPV only infects the Muscovy duck, while GPV can infect both geese and Muscovy ducks. MDGPV is pathogenic to geese and Muscovy ducks in artificial infection tests. However, MDGPV and MDPV NS genes possessed high sequence homology, and the recombination of genes should be further studied.
NS proteins in members of the Parvovirus family play an important role in viral replication, transcription, and infection of host cells, associated with viral virulence [15-21]. At present, the nucleoside triphosphate (NTP)-binding motif in the NS protein structure is considered to be the principal region producing toxicity. Amino acid mutations in this region affect the function of Walker A, the structural unit of the ATPase active region of helicase superfamily members, which alters the cytotoxicity of the virus towards the host. Using multisequence alignment analysis,it was found that the differences in strong and weak MDGPV strains at the amino acid site aa338 in the NTP region were similar to the viral attenuated effect caused by the mutation of the parvoviral NS gene [22- 24]. Furthermore, this mutation caused a disappearance in an MCH II CD4+ T-cell epitope (aa333-aa341) from the parental virulent PT strain. However, the B-cell epitope functionality was preserved, which can be inferred from the difference in amino acid residues,a determinant of MDGPV toxicity.
Currently, study of GPV and MDPV genome characteristics focuses on analysis of genetic variation in viral structural protein genes. It is believed that there is a high degree of conservation in gene sequences between GPV and MDPV strains of different ages in the same region, and even between the strong parental virus and that which is attenuated. The two functional open reading frames between GPV and MDPV have also high homology. MDGPV showed nucleotide sequence characteristics of MDPV on the NS gene, indicating that GPV is still actively involved in the immune stress of the host. The study of NS protein function of MDGPV is the basis for revealing the pathogenesis of the gosling disease and to identify the strong and weak strains. The intensive use of bioinformatics software provided a deeper and better understanding of the biological information contained in MDGPV, and also provided evidence relating to the molecular biology that explained the origins of MDGPV.
References
1. Yang J, Yang R, Cheng A, Wang M, Fu L, Yang S, Zhang S, Yang L, Xu Z. A simple and rapid method for detection of Goose Parvovirus in the field by loop-mediated isothermal amplification. Virol J 2010; 7:14
2. Wang S, Cheng XX, Chen SY, Zhu XL, Chen SL, Lin FQ, Li ZL. Genetic characterization of a potentially novel goose parvovirus circulating in Muscovy duck flocks in Fujian Province, China. J Vet Med Sci 2013;75:1127-1130.
3. Liu HM, Wang H, Tian XJ, Zhang S, Zhou XH, Qi KZ, Pan L. Complete genome sequence of goose parvovirus Y strain isolated from Muscovy ducks in China. Virus Genes 2014;48:199-202.
4. Zádori Z, Stefancsik R, Rauch T, Kisary J. Analysis of the complete nucleotide sequences of goose and muscovy duck parvoviruses indicates common ancestral origin with adeno- associated virus 2. Virology 1995;212:562-573.
5. Niu Y, Zhao L, Liu B, Liu J, Yang F, Yin H, Huo H, Chen H. Comparative genetic analysis and pathological characteristics of goose parvovirus isolated in Heilongjiang, China. Virol J 2018;15:27.
6. Wang CY, Shieh HK, Shien JH, Ko CY, Chang PC. Expression of capsid proteins and non-structural proteins of waterfowl parvoviruses in Escherichia coli and their use in serological assays. Avian Pathol 2005;34:376-382.
7. Wang J, Duan J, Zhu L, Jiang Z, Zhu G. Sequencing and generation of an infectious clone of the pathogenic goose parvovirus strain LH. Arch Virol 2015;160:711-718.
8. Nüesch JP, Tattersall P. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology 1993;196:637-651.
9. Shackelton LA, Hoelzer K, Parrish CR, Holmes EC. Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol 2007;88:3294-3301.
10. Shen H, Zhang W, Wang H, Zhou Y, Shao S. Identification of recombination between Muscovy duck parvovirus and goose parvovirus structural protein genes. Arch Virol 2015;160:2617- 2621.
11. Wang S, Cheng X, Chen S, Lin F, Chen S, Zhu X, Wang J. Evidence for natural recombination in the capsid gene VP2 of Taiwanese goose parvovirus. Arch Virol 2015;160:2111-2115.
12. Wang S, Cheng XX, Chen SY, Lin FQ, Chen SL, Zhu XL, Wang JX, Huang MQ, Zheng M. Phylogenetic analysis of VP1 gene sequences of waterfowl parvoviruses from the Mainland of China revealed genetic diversity and recombination. Gene 2016;578:124- 131.
13. Wang J, Ling J, Wang Z, Huang Y, Zhu J, Zhu G. Molecular characterization of a novel Muscovy duck parvovirus isolate: evidence of recombination between classical MDPV and goose parvovirus strains. BMC Vet Res 2017;13:327.
14. Li P, Lin S, Zhang R, Chen J, Sun D, Lan J, Song S, Xie Z, Jiang S. Isolation and characterization of novel goose parvovirus- related virus reveal the evolution of waterfowl parvovirus. Transbound Emerg Dis 2018;65:e284-e295.
15. Chang PC, Shien JH, Wang MS, Shieh HK. Phylogenetic analysis of parvoviruses isolated in Taiwan from ducks and geese. Avian Pathol 2000;29:45-49.
16. Yan YQ, He TQ, Li R, Zhang SY, Wang K, Yi SS, Niu JT, Dong H, Hu GX. Molecular Characterization and Comparative Pathogenicity of Goose Parvovirus Isolated from Jilin Province, Northeast China. Avian Dis 2019;63:481-485.
17. Li D, Zhang L, Chen S, Gu J, Ding M, Li J. Detection and Molecular Characterization of Two Genotypes of Goose Parvoviruses Isolated from Growing Period Geese and Cherry Valley Ducks in China. Avian Dis 2019;63:411-419.
18. Wang J, Duan J, Meng X, Gong J, Jiang Z, Zhu G. Cloning of the genome of a goose parvovirus vaccine strain SYG61v and rescue of infectious virions from recombinant plasmid in embryonated goose eggs. J Virol Methods 2014;200:41-46.
19. Shien JH, Wang YS, Chen CH, Shieh HK, Hu CC, Chang PC. Identification of sequence changes in live attenuated goose parvovirus vaccine strains developed in Asia and Europe. Avian Pathol 2008;37:499-505.
20. Tsai HJ, Tseng CH, Chang PC, Mei K, Wang SC. Genetic variation of viral protein 1 genes of field strains of waterfowl parvoviruses and their attenuated derivatives. Avian Dis 2004;48:512-521.
21. Tatár-Kis T, Mató T, Markos B, Palya V. Phylogenetic analysis of Hungarian goose parvovirus isolates and vaccine strains. Avian Pathol 2004;33:438-444.
22. Jindal HK, Yong CB, Wilson GM, Tam P, Astell CR. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J Biol Chem 1994;269:3283-3289.
23. Li G, Sun C, Zhang J, He Y, Chen H, Kong J, Huang G, Chen K, Yao Q. Characterization of Bombyx mori parvo-like virus non-structural protein NS1. Virus Genes 2009;39:396-402.
24. Pécsi I, Szabó JE, Adams SD, Simon I, Sellers JR, Vértessy BG, Tóth J. Nucleotide pyrophosphatase employs a P-loop-like motif to enhance catalytic power and NDP/NTP discrimination. Proc Natl Acad Sci U S A 2011;108:14437-14442.






















